

Gene Lab
GeneLab is a multi-faceted core laboratory directed by the Division of BIOMMED in the School of Veterinary Medicine at Louisiana State University. GeneLab engages in specific research and training projects, which require expertise in Next-Generation Sequencing, traditional DNA sequencing, gene cloning, PCR, gene expression and other molecular methods. The goal of GeneLab is to facilitate the utilization of the state-of-the-art technologies in genomics research by LSU faculty and researchers nationwide at a competitive price and in a timely fashion. All GeneLab services can be accessed through iLab . Login to your iLab
The primary focus of GeneLab is its portfolio of sequencing capabilities. Currently, two Next Generation Sequencing instruments, the Ion Torrent PGM and the Ion Proton along with bioinformatics support for NGS data are provided to the research community and our offerring will be extended rapidly as NGS and other emerging sequencing technologies are evolving.
GeneLab also offers a range of traditional sequencing and fragment analysis options using our ABI 3130 Genetic Analyzer.
Researchers utilizing GeneLab have access to various services and user-driven instrumentation. See Equipment List for specific instruments available.
GeneLab also operates a Supply Center offering scientific reagents and supplies from Life Technologies (formerly Applied Biosystems and Invitrogen Corp) and oligonucleotides from IDT (Integrated DNA Technologies). Discounted pricing and free shipping have been negotiated with these vendors to ensure that LSU researchers get the most value for their research dollar.
Since its inception in 1990, GeneLab has processed more than 15,000 samples and served approximately 75 Louisiana state laboratories and numerous out of state research institutions.
Acknowledgments for Use of GeneLab equipment:
Welcome to iLab Solutions
iLab Solutions provides web-based services designed to streamline research operations. GENELAB uses iLab’s Core Management Platform to support reservations, service requests (including training and purchasing), equipment tracking, project management automated billing, and reporting
Firefox is preferred when using the iLab application. Safari or Chrome are also acceptable browser options. At present Internet Explorer does not behave consistently for some iLab functionality depending on the version. iLab is addressing this.
iLab Account
To register for a new account:Contact:
Thaya StouffletCore Manager
Office: SVM 3212B
E-mail: thaya@lsu.edu
Phone: (225) 578-9708
Login to your existing account:
iLab Chat Tool - click on the chat link or popup Help
Login to iLab and use the online support portal
Email your questions to: support@ilabsolutions.com
Call iLab Support at :617.297.2805 (Eastern time business hours)
iLab support page: Support
iLab FAQ page: FAQ's
| Sample Preparation | |
| gDNA library preparation (Ion PGM/Proton) | $150 |
| cDNA library for the RNA-Seg(Ion PGM/Proton) | $250 |
| gDNA library preparation (Ion Proton) | $150 |
| ChIP-Seq library preparation (Ion PGM) | $150 |
| Sequencing with PGM | |
| DNA sequencing using 314 Chip | $600 ($750 if library prep is included) |
| DNA sequencing using 316 Chip | $800 ($950 if library prep is included) |
| DNA sequencing using 318 Chip | $900 ($1050 if library prep is included*) |
| cDNA sequencing using 316 Chip | $800 ($1050 if library prep is included*) |
| cDNA sequencing using 318 Chip | $900 ($1150 if library prep is included*) |
| ChIP-Seq sequencing using 316 Chip | $900 ($1150 if library prep is included*) |
| ChIP-Seq sequencing using 318 Chip | $900 ($1050 if library prep is included) |
| Sequencing with Proton (Ion proton) | |
| DNA sequencing using Ion PI Chip | $1300 ($1450 if library prep is included) |
| cDNA sequencing using Ion PI Chip | $1300($1550 if library prep is included*) |
| RNA-Seq data analysis | |
| Read alignment/Expression Estimation/Differential Gene Expression | $480 (2 hours-interval) Service limited to GeneLab NGS |
| ChIP-Seq data analysis | |
| Read alignment/Peak Calling | $480 (2 hours-interval) service limited to GeneLab NGS data |
| Other data analysis | |
| Quality control/Alignment | $480 (2 hours interval) service limited to GeneLab NGS data |
| SNP and small lndel finding | $480 (2 hours interval) service limited to GeneLab NGS data |
| Assembly | $480 (2 hours interval) service limited to GeneLab NGS data |
| Advanced consulting for RNA- Seq data | Request-based |
| BigDye Terminator Reaction, Dye Removal, Electrophoresis, Analysis (per sample) | $11.00 |
| BigDye Terminator Reaction, Dye Removal, Electrophoresis, Analysis (96well plate)**96 well plate must be provided preloaded with samples and primers | $816.00 |
| Electrophoresis Only | $4.00 |
| Fragment Analysis | $8.00 |
| Sequence Electropherogram Printout (per sample) | $1.00 |
| Pre-sequencing processing/clean-up from PCR product/agarose gel slice | $5.00 |
| Sequence Primer Walking (per 400 bases) | $25.00 |
| Real-time PCR (7500/7900/Light Cycler) Run | $15.00 |
| Real-time PCR (7500/7900/Light Cycler)Technician Assisted DNA Run/Reagents | $250.00 |
| Real-time PCR (7500/7900/Light Cycler)Technician Assisted RNA Run/Reagents | $350.00 |
| BioPlex Usage (per run) | $200.00 |
| Accuri C6 Flow Cytometer - Daily Usage Fee | $25.00 |
| Accuri C6 Flow Cytometer - Hourly Rate (in addition to daily usage fee) | $25.00 |
| Invitrogen Countess Automated Cell Counter Usage Fee | $1.00 |
| Nexcelom Cellometer Vision Automated Cell Counter Usage Fee | $1.00 |
| Delivery/Pick-up fee | $5.00 |
| Alpha Innotech Fluorochem Q imaging station | |
| Bio-Rad GenePulser Xcell Electroporation System | |
| SpectraMax M2 Plate Reader | |
| Turner Designs TD-20/20 Luminometer | |
| Agilent 2100 Bioanalyzer | |
| NanoDrop ND-1000 Spectrophotometer |

GeneLab now operates a Supply Center offering scientific reagents and supplies from the two divisions of Life Technologies, Invitrogen, Inc. and Applied Biosystems, Inc. Discounted pricing has been negotiated with these vendors to ensure that LSU researchers get the most value for their money. In addition, free shipping will be offered to all orders shipped directly to GeneLab. For most orders this is an additional savings of 15-25%. Investigators have the cost-free option of picking up their orders from GeneLab (located in room 3212 of the LSU School of Veterinary Medicine) or having their orders delivered to an LSU campus address for an additional $5 to be invoiced via an LSU Internal Transaction.
Supply Center Pick-up Hours: 8:00am-11:30am Monday-Friday (or by appointment)
Supply Center Delivery Hours: 2:00pm-4:00pm Tuesday, Thursday, and Friday

Invitrogen offers 35,000 unique products and services to support disease research, drug discovery and commercial bio-production. Invitrogen’s product family includes many of the most widely recommended names in the industry, including Invitrogen, Molecular Probes, Gibco, BioSource and Dynal Biotech.
The Invitrogen portion of the GeneLab Supply Center will be offered through a Supply Center Management System (SCMS) hosted directly at Invitrogen's web server. Discounted pricing has been negotiated for all orders placed through the SCMS system and and free shipping to those shipped directly to GeneLab. (Investigators have the cost-free option of picking up their orders from GeneLab (located in room 3212 of the LSU School of Veterinary Medicine) or having their orders delivered to an LSU campus address for an additional $5 to be invoiced via an LSU Internal Transaction.) Before placing an order, individuals must register at the Invitrogen website (Registration Link) where he/she will receive a username and password via email. During this registration process, be sure to note 'LSU GeneLab' in Supply Center location box. Once you have received your username/password via email (usually 2 business days) you can login to the SCMS system (Login)

Applied Biosystems is a leading brand of scientific reagents, services, supplies and equipment that aid researchers in Cell Biology, DNA Sequencing, Gene Expression, Genotyping, Mass Spectrometry, Microarrays, PCR/RT-PCR, Protein Sequencing, and Real-Time PCR among others.
The Applied Biosystems portion of the GeneLab Supply Center will be offered directly through GeneLab. LSU Investigators will be invoiced via LSU Internal Transcation. All orders will be placed via an online PDF form (GeneLab-ABI Order Form) sent directly to GeneLab. Any items not stocked in GeneLab itself will be special ordered as needed. Discounted pricing and free shipping to GeneLab will be offered to all items ordered through the Supply Center. Investigators have the cost-free option of picking up their orders from GeneLab (located in room 3212 of the LSU School of Veterinary Medicine) or having their orders delivered to an LSU campus address for an additional $5 to be invoiced via an LSU Internal Transaction.
To place an Applied Biosystems order:(Requires Adobe Reader 7.0 or later)
- Complete the GeneLab-ABI order form completely.
- Save a copy of the form to your desktop.
- Email the form to genelab@lsu.edu
- You will receive an order confirmation email shortly.
- You will receive an email when your product is ready for pick-up or delivery.
Questions on Registrations?
Phone: 888.584.8840
supplycenters@invitrogen.com



LSU GeneLab, in partnership with Integrated DNA Technologies (IDT), now offers custom oligonucleotides and other molecular biology services. Benefits to the researcher using this service include top-quality molecular tools, reduced pricing and free shipping if items are picked up in GeneLab (SVM 3212). Delivery to your lab can be arranged for a fee of $5 per delivery. Orders are placed using a simple web-based system. See instructions below for setting up an account and ordering.
To set up an account
- Go to the GeneLab Primer Service portal.
- Click on the setup New Account†button.
- Complete online form.
- Click “Saveâ€.
- You will be redirected to the order page.
- You can place the order now, following the ordering instructions beginning with step 2.
To place an order
- Login.
- Choose the type of service or product you need from the Products page.
- Complete the order form following on-line instructions.
- When you have entered all of your order information, click Add to Order button.
- Verify your information is correct.
- Click Checkout.
- Enter only your phone number in the Shipping section.
- Click Continue.
- Select Documentation Option of your choice if prompted.
- Click Continue.
- Enter your departmental account number followed by your Principal Investigator’s 3 initials and four zeroes, separated by a backslash. For example: (123-45-6789/ABC0000)
- Click Submit.
- Click Logout.
An email confirmation will follow shortly informing you that your order is awaiting approval from GeneLab.  After the order is approved and released by GeneLab, you will receive an email confirmation that your order has been placed. Your order will be shipped to GeneLab for pick-up with no shipping charges. If you wish to have GeneLab deliver the order to your lab, an additional $5.00 will be charged to the account provided in the form of an internal transaction (IT).
If you have any questions, please contact GeneLab at genelab@lsu.edu
GeneLab launched its NGS and bioinformatics support services initially in January, 2013 under a developmental stage and officially in July 1, 2013 to assist researchers at all stages of research, from experimental design to data generation and analysis of NGS data. The close alignment of GeneLab’s NGS capabilities and bionformatics provides an integrated approach to experimental design, NGS experimentation and fast analysis of the project data, while creating a unique forum for cross-training biologists and computer scientists in NGS methodologies and bionformatics. GeneLab is equipped with a suite of Life Technologies NGS equipment including:
- Ion Torrent PGM .
- Quant Studio Flex (digital PCR validation) .
- Ion Proton .

Both the Ion Torrent PGM and Ion Proton have web-based Ion Torrent server with which produced sequencing data access and integrated data analysis pipelines are available. GeneLab-Bioinformatics Unit operates two high-end servers. The visualization server supports windows as well as linux environments, providing memory intensive visualization tasks and tools working on the two OS environments. A different, high-end server is equipped with large memory (128 GB) and large number of cores (32), is networked to the Ion PGM and Ion Proton platforms for dealing with analyses requiring large memory footprint and multi-core performance applications. These two high-end servers are backed with network-connected expandable NAS storage system, resulting in total storage space of 90 Terabytes at this time, which are directly connected to the servers and software tools integrated to the sequencing platforms. NextGen sequencing and bioinformatics analysis services can be scheduled via iLab.
GeneLab, also offers capillary DNA sequencing services for a wide range of templates including plasmid DNA, PCR fragments, cosmid DNA, bacterial genomic DNA, BAC and cDNA. Clients who have large inserts can take advantage of our primer walking service (see below) to get information on their entire clone.
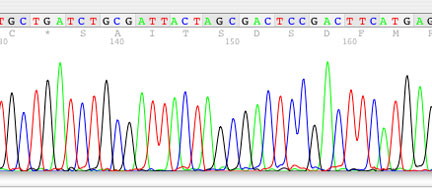
We perform standard sequencing reactions using Applied BioSystems BigDye Terminator version 3.1 or version 1.1 (depending on template) chemistries. Reactions are run on either of our two ABI Prism 3130, a four capillary based DNA sequencer.
Sample Submission Requirements
- No DNA/Primer premixing
- No special sample tubes
- Quantitate your DNA
- The #1 reason sequencing reactions fail is that researchers do not quantitate their samples. Please quantitate your DNA by OD, agarose gel, or the NanoDrop -1000 available in GeneLab.
- Send DNA and Primers separately
- The main benefits of NOT premixing include: Additional reactions may be performed on a particular DNA template without having to provide more sample AND GeneLab can troubleshoot projects by adjusting DNA-primer concentrations..
- You will be redirected to the order page.
- You can place the order now, following the ordering instructions beginning with step 2.
- Send template sample(s) and primer(s) to GeneLab. Please see Template Concentrations below for appropriate amounts of template to submit. All samples must be submitted through iLab.
- GeneLab provides standard sequencing primers (see below) at no charge. If using your own primers, please see Primer Concentrations (above) and include with template shipment.
| Template | Concentration | Volume |
| Plasmid | >20ng/µL | 10 µL |
| PCR < 500bp | 2ng/ µL | 10 µL |
| PCR>500bp | >2ng/ µL | 10 µL |
| Cosmid | 0.5-1.0 μg | 10 µL |
| BAC DNA | 0.5-1.0 μg | 10 -µL |
| Primer | 2 µM | 10 µL |
*We typically request more DNA than we need in order to have a sufficient quantity for any repeats that may need to be performed. A general rule of thumb for PCR products is to submit 2 ng/µl per 100 bp.
Universal primers provided at no extra cost (see table below)
| Primer Name | Primer Sequence |
| M13 Forward (-20) | GTA AAA CGA CGG CCA GT |
| M13 Forward (-40) | GTT TTC CCA GTC ACG AC |
| M13 Reverse (-24) | AAC AGC TAT GAC CAT G |
| M13 Reverse (-48) | AGC GGA TAA CAA TTT CAC ACA GGA |
| T7-19 | GTA ATA CGA CTC ACT ATA G |
| T7-Rev | GCTAGTTATTGCTCAGCGG |
| T3-19 | CAA TTA ACC CTC ACT AAA G |
| KS-18 | TCG AGG TCG ACG GTA TCG |
| SK-20 | CGC TCT AGA ACT AGT GGA T C |
| SP6 | GAT TTA GGT GAC ACT ATA G |
GeneLab uses the Sequencing Cycle Run Parameters below on a PTC 200 Thermalcycler from MJ Research:
40 cycles of:
- 96° for 10 seconds
- 50° for 5 seconds
- 60° for 4 minutes
In general, a single run should result in at least 500-700 bases of high quality sequence information. Turn-around time is generally 1-2 days from receipt of the template and primers, depending on the workload at the facility. Investigators will receive the results as a sequence file and a color electropherogram for each template/primer combination. Files will be sent by e-mail to all investigators. For more information regarding automated DNA sequencing, see Applied Biosystems' Sequencing Chemistry Guide for Automated Sequencing. To visualize and print the chromatogram traces, please point your favorite web browser to the following URLs to obtain free software for Windows based computers
CodonCode Aligner (for academic users only)
Sequencing by primer walking
This is an effective strategy for the efficient sequence determination of cloned DNA fragments which range in size from 0.5-8 kb. Initially sequence data is obtained using a standard primer which hybridizes to the vector sequence upstream of the insert DNA. Based on this initial data, GeneLab will design a custom oligonucleotide, have it synthesized, and use it to prime a second sequencing reaction. The data obtained from the second reaction should overlap with the initial data and extend the sequence further downstream of the initial primer. By repeated cycles of custom oligonucleotide synthesis and DNA sequencing, the cloned insert is sequenced completely in one direction. Pricing is dependent upon size of the project.
For DNA sequencing rates, please see the " GeneLab Rates page "
For more info contact:
Thaya Guedry
Lab Manager
Phone: 225.578.9708
genelab@lsu.edu

Combining flexible throughput capabilities with a streamlined workflow, the QuantStudio™ 12K Flex system takes you from targeted discovery through confirmation and screening, all on a single platform.
- Five interchangeable blocks: (OpenArray® plate, TaqMan® Array Card, 384-well, Fast 96-well, and 96-well blocks).
- Provides a seamless switch from qPCR to digital PCR using QuantStudio™ Digital PCR kits and DigitalSuite Software with the QuantStudio™ OpenArray® block.
- Maximum multiplexing & chemistry options with Enhanced OptiFlex® System.
- Comprehensive software analysis tools for gene expression, genotyping, and digital PCR.
GeneLab also maintains four Real-Time PCR instruments: one Roche LightCycler 480 II, two ABI 7900 Sequence Detection Systems. All are in room 3212 of the LSU School of Veterinary Medicine.

The Roche LightCycler 480 Real-Time PCR System is a fully integrated multiwell-plate based real-time PCR platform for highly accurate qualitative and quantitative detection of nucleic acids. Building on the benefits of Roche's capillary-based LightCycler Systems, it goes one step further in offering enhanced throughput, compatibility with automation equipment and maximum flexibility regarding hard- and software. Providing novel ways to combine speed and accuracy without compromises, the LightCycler 480 Real-Time PCR System meets the needs of a broad range of applications in research fields such as gene expression studies, discovery and analysis of genetic variation or array data validation.
- Achieve faster and easier assay setup with the new sample editor.
- Set up experiments with just a few clicks, with options to refine your results later.
- Benefit from enhanced templates and macros for all applications.
- Perform genotyping based on endpoint analysis or melting curves.
- Use basic and advanced methods for gene expression and genotyping.

The Applied Biosystems 7900HT Fast Real-Time PCR System is the only real-time quantitative PCR system that combines 384-well plate compatibility with fully automated robotic loading and now also offers optional Fast real-time PCR capability.
- Fast PCR option reduces run time to about 35 minutes in a standard 96-well format, or about 55 minutes in a 384-well plate.
- Continuous wavelength detection from 500-660 nm allows the use of multiple fluorophores in a single reaction.
- Measure gene expression levels, detect and quantitate pathogens, perform allelic discrimination (SNP genotyping) assays as well as score the presence of gene sequences.
- 96- or 384-well plate compatibility (including the TaqMan Low Density Array).
- Optional Enterprise edition software provides data analysis tools that support 21 CFR Part 11 guidelines providing data integrity and security.
- Hands-free plate-loading and unloading provides true walkaway automation allowing you to increase your lab's productivity.
- Proven assay development guidelines save time and money
Applied BioSystems has developed two types of chemistries to detect PCR products using Sequence Detection Systems (SDS) instruments:
- TaqMan chemistry (also known as fluorogenic 5' nuclease chemistry)
- SYBR Green I dye chemistry
Genelab staff offers advice and training for users of these instruments. Contact Thaya Guedry for more information on how to schedule a run through the
For more info contact:
Thaya Guedry
Lab Manager
Phone: 225.578.9708
genelab@lsu.edu
GeneLab houses an Accuri C6 Flow Cytometer, Nexcelom Biosciences Cellometer Vision Cell Profiler, and an Invitrogen Countess automated cell counter.
C6 Flow Cytometer

The Accuri C6 Flow Cytometer, CFlow Software and Workstation PC supply all the capabilities of a full-featured flow cytometer. The C6 system includes blue and red lasers, four color detectors and both forward and side scatter detectors plus software that is extremely intuitive.
Advanced Features:
- Over 7 Decade dynamic range
- Optical bench needs no alignment
- Fluidics control
- Absolute counts
- Maintenance made simple
- Works with current protocols and reagents
- FCS 3.0 compatible file format
- Compact size
- Low power usage
C6 Specifications
Cellometer Vision

The Cellometer Vision Cell Profiler combines brightfield microscopy and multi-channel fluorescence images to generate cell count and fluorescence data. It is most suitable for determining cell concentration and viability in complex samples. The sophisticated data analysis software allows detailed studies of select cell population. Cellometer Vision enables high sensitivity fluorescence detection, suitable for counting cells with reporter proteins such as GFP or RFP. Cellometer Vision is also used for measuring cells with fluorescence-conjugated antibodies.
Best Suited For:
- Total cell concentration for highly viable cell population
- Live cell concentration and viability using trypan blue
- Cell size measurement
- Identify and count cells based on cell size difference
- Capture and store cell images
- Total nucleated cell concentration without lysing non-nucleated cells
- Live nucleated cell concentration without lysing non-nucleated cells
- Fluorescence based live cell concentration and viability for cell lines
- Transfection efficiency by GFP
- Transfection efficiency by YFP
- Transfection efficiency by RFP
- Live cell concentration and viability using two fluorescents in one sample
- AO, PI, EB, DAPI, Hoechst
Countess Cell Counter

The Countess provides fast, easy and accurate cell counting without using a hemocytometer, eliminating the tedium and subjectivity of manual cell counting forever. The Countess Automated Cell Counter uses trypan blue staining combined with a sophisticated image analysis algorithm to produce accurate cell and viability counts in just 30 seconds. The algorithm also measures average cell size of live, dead, and total cells to give you all the data you need to proceed with your experiments. The measurement range extends from 1 x 104 to 1 x 107 cells/mL, with an optimal range from 1 x 105 to 4 x 106 cells/mL, broader than that of a hemocytometer. You can save data to a user-provided USB drive for later analysis. Free Countess software is available to download from Invitrogen's website.
- Eliminates subjectivity of manual counting
- Minimizes human error
- Provides reproducible data
- Counts live and dead cells
- Provides viability
- Measures average cell size
- No setup or maintenance
- Uses just 10μL of sample
- Includes dilution calculator and data archiving
Countess Automated Cell Counter Product Manual
For more info contact:
Thaya Guedry
Lab Manager
Phone: 225.578.9708
genelab@lsu.edu

The BioRad BioPlex uses Luminex technology to analyze microsphere-based multiplex protein assays. This is an alternative to using ELISA assays to determine the concentration of proteins in solution, such as cell lysates. The advantage of doing multiplex assays is that one can get results simultaneously for many proteins dissolved in a very small volume. It can perform analysis of up to 100 different biomolecules (proteins, peptides, or nucleic acids) in a single microplate well. The microplate platform allows the automated analysis of 96-well plates with a throughput of more than 1,800 assay points (19plex assay) in 30 minutes.
Using the BioPlex software, standard curves of known concentration of all analytes are automatically computed and data from the standards and unknowns can be displayed in graphic or table form. The results can also be saved in either form, whichever is preferred by the investigator.
Kits for simultaneous quantitative measurement of up to 25-30 proteins are available. Examples of proteins that can be analyzed with commercially prepared Luminex microspheres are human/murine/rat cytokines, human/murine phosphoproteins, growth factors, kinases, and transcription factors. Users may also create custom assays for any proteins having corresponding antibodies.
Keep in mind that it takes several hours to set up the plates for the BioPlex, so it is helpful to start early in the morning. You may prepare your plates in room 3101 of the LSU School of Veterinary Medicine if you do not have a vacuum manifold in your laboratory.
Visit http://www.bio-rad.com for more on the BioPlex.
For more info contact:
Thaya Guedry
Lab Manager
Phone: 225.578.9708
genelab@lsu.edu

Knowing how much the quality of RNA affects the success of dye labeling and hybridization to microarray slides along with various other applications, GeneLab offers the use of an Agilent 2100 Bioanalyzer. Housed in Room 3212 of the School of Veterinary Medicine , the Bioanalyzer runs "micro gels" of both total and poly-A RNA and shows an electropherogram image and gel-like image of the sample. It gives the 28s/18s peak ratio, percent of DNA contamination and the approximate RNA concentration. The images help to visualize any RNA degradation and provide a quantitative value for the peak ratio on total RNA samples. In addition to RNA, the Bioanalyzer can be used to determine the quality of both DNA and protein.
The Bioanalyzer holds one chip with room for up to 12 samples, a combination of 1ul of 100ng/ul RNA and 5uls of a gel-dye mix. These samples are forced through micro-channels to perform electrophoretic separation, and then are forced through a final channel where a laser is located to detect the segments as they pass. The samples are compared with a standard ladder also run on the same chip. All 12 samples' gel-like images are aligned by a marker in the dye mix. One chip takes less than 30 minutes to complete, and digital data is available as each sample is analyzed for immediate results.
Results from the Bioanalyzer are highly accurate and repeatable, and have become a necessary procedure before submitted RNA is used for other experiments
Keep in mind that it takes several hours to set up the plates for the BioPlex, so it is helpful to start early in the morning. You may prepare your plates in room 3101 of the LSU School of Veterinary Medicine if you do not have a vacuum manifold in your laboratory.
Visit http://www.bio-rad.com for more on the Agilent 2100 Bioanalyzer.
For more info contact:
Thaya Guedry
Lab Manager
Phone: 225.578.9708
genelab@lsu.edu

GeneLab offers the use of the FluorChem Q from Cell Biosciences and AlphaInnotech to all researchers. The FluorChem Q is an imaging station with the capability to image ethidium bromide gels, chemifluorescent gels, and chemiluminescent membranes such as dot blots. Using UV light, white light and various filters, the FluorChem Q is a versatile instrument, yet remains user-friendly. The multiplex fluorescent detection and chemiluminescent imaging capabilities of the FluorChem Q provide a complete solution for quantitative Western blot imaging and analysis. The open platform is compatible with a wide variety of dyes and kits, providing the flexibility needed for multiple applications.
- Six-position filter wheel provides UV and fluorescent imaging versatility
- Three excitation channelswith specific wavelengths for multiplex fluorescent Western blot imaging
- Powerful fixed lens for rapid imaging speeds
- AlphaView® Q software saves imaging protocols and provides quantitative analysis tools
There is no charge for images at the present time. Images can be printed on thermal paper and/or saved to data storage devices such as CD's or portable USB drives. GeneLab staff can offer assistance and training in the use of the equipment. Stand-alone software is available to allow the researcher to edit images in his own laboratory.
For more info contact:
Thaya Guedry
Lab Manager
Phone: 225.578.9708
genelab@lsu.edu
Have to update
Contact Us
Have to update.
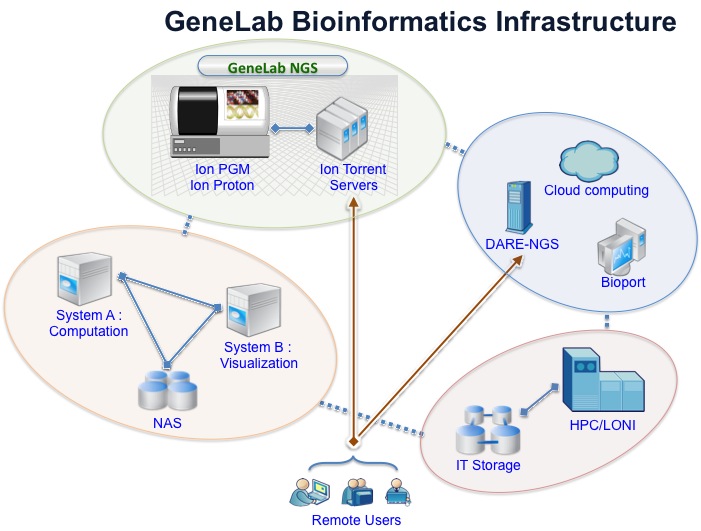
Bioinformatics Support Overview
GeneLab offers Next Generation Sequencing (NGS) data analytics with a broad range of programs such as tools of Ion Torrent suite, tools from Life technologies, community-wide recognized bioinformatics tools, and commercial desktop tools for NGS sequencing and downstream analsyis. Our data analytics tasks are facilitated by bioinformatics infrastructure that comprises backend computing resources including high-end servers, network storage and HPCs, software tools, and various in-house services provided by DARE-based science gateways and a local blast infrastructure. The right schematic illustrates our bioinformatics infrastructure supporting our service that utilizes avaialble tools.
Data analyses for NGS inlcude- RNA-Seq data analysis using our pipelines
- RNA-Seq data analysis using commercial packages including Partek (workflow, genome suite, and pathway)
- Various analyses such as alignment, variant calling, de novo assembly, and many from Ion Torrent plug-ins
- Analsyis using standalone tools for alignment, de novo assembly, RNA-Seq., ChIP-Seq., and others
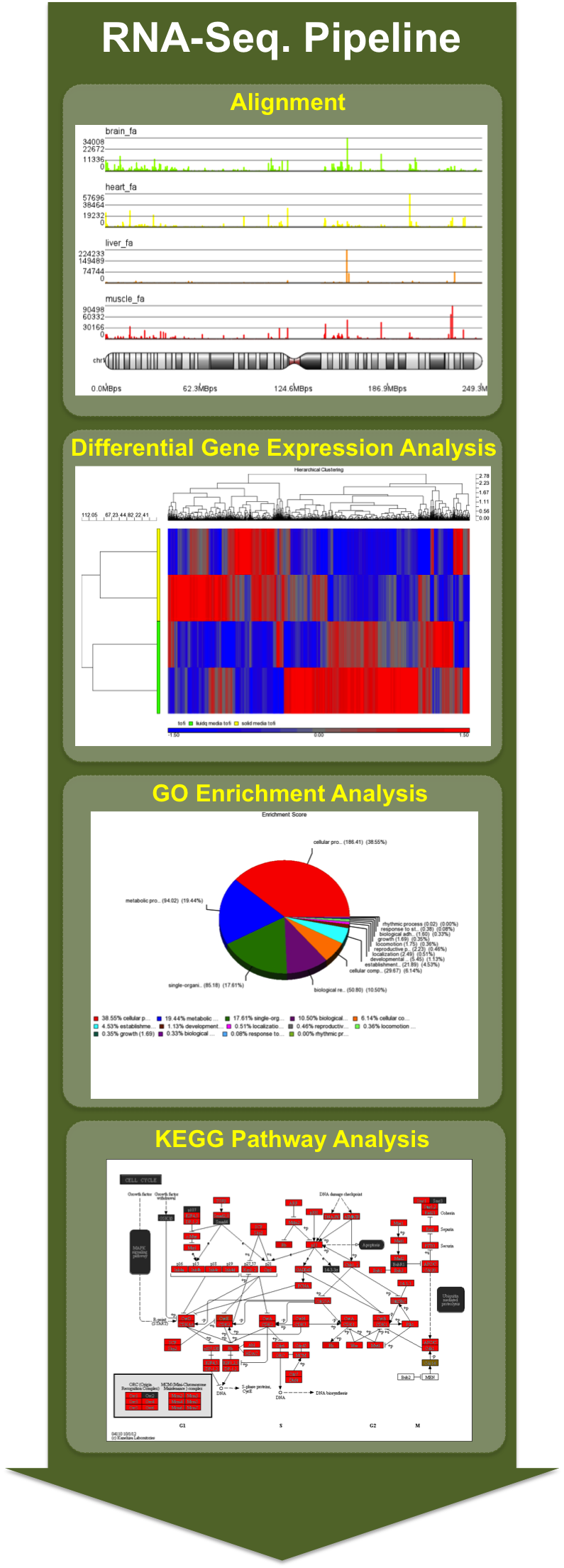
RNA-Seq Pipline
Exome Pipline
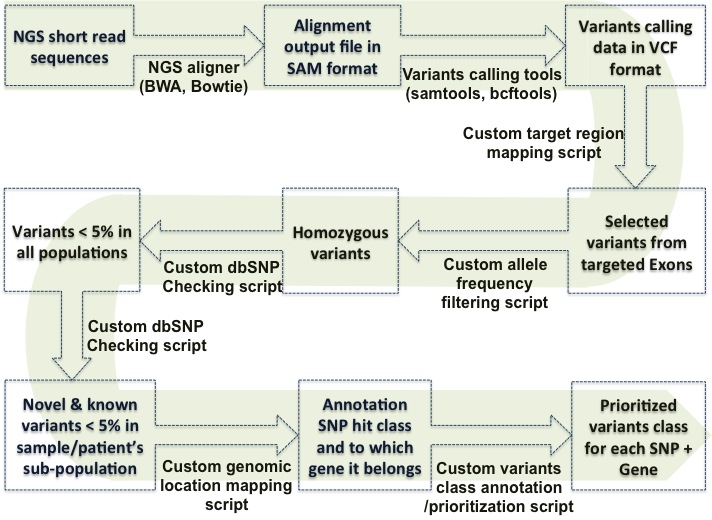
Supporting software tools (Examples)
| Pipelines | RNA-Seq., ChIP-Seq. |
| Alignment | BWA, Bowtie, Bowtie2, BFAST, Mosaic, RUM |
| De novo asembly | ABySS, MIRA3, Velvet |
| Variant calling | SAMTools, GATK |
| Misc | fastX-toolkit, fastQC, R/Bioconductor, Galaxy, Local Blast service |
| Commerical packages | DNASTAR, Partek |
| Genome browsing | Artemis, IGV, GBrowse |
Bioinformatics Support Team:
Joohyun Kim, PhD
jhkim@cct.lsu.edu
Nayong Kim, PhD
nykim@cct.lsu.edu
Shayan Shams
Graduate Assistant
sshams2@cct.lsu.edu
